11. Exercícios de treino, 2#
11.1. Objetivo#
Estes exercícios ilustram o uso dos módulos math, numpy e matplotlib.
Nos programas seguintes, preencha os '??????' com expressões apropriadas de forma a garantir que o resultado da execução do programa seja aquele que está exibido
11.2. Numéricos, sobre comandos for, listas em compreensão e numpy#
11.2.1. Programa 1#
import math
from matplotlib import pyplot as plt
fibs = [0, 1, 1, 2, 3, 5, 8, 13, 21, 34, 55, 89, 144, 233, 377, 610, 987, 1597, 2584, 4181, 6765, 10946, 17711, 28657]
# logaritmos dos números com math.log()
# mas não pode dar erro...
logfibs = '??????'
plt.plot(range(len(logfibs)), logfibs, 'o-')
plt.show()
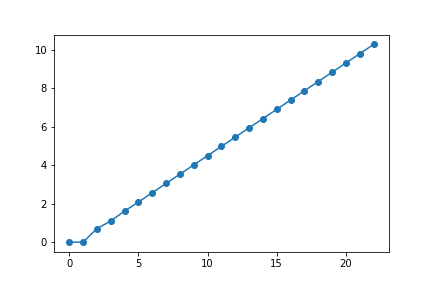
Solução
Show code cell content
import math
from matplotlib import pyplot as plt
fibs = [0, 1, 1, 2, 3, 5, 8, 13, 21, 34, 55, 89, 144, 233, 377, 610, 987, 1597, 2584, 4181, 6765, 10946, 17711, 28657]
# logaritmos dos números com math.log()
# mas não pode dar erro...
logfibs = [math.log(x) for x in fibs if x != 0]
plt.plot(range(len(logfibs)), logfibs, 'o-')
plt.show()
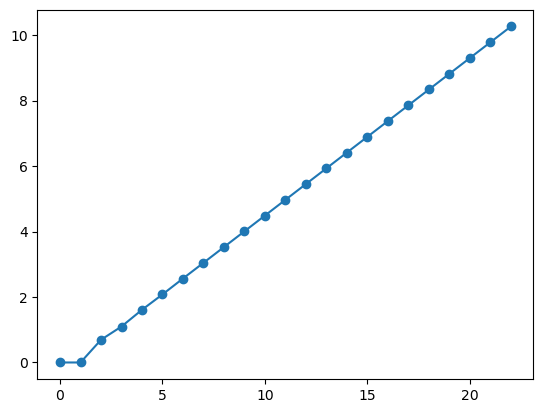
11.2.2. Programa 2#
import numpy as np
from matplotlib import pyplot as plt
fibs = [0, 1, 1, 2, 3, 5, 8, 13, 21, 34, 55, 89, 144, 233, 377, 610, 987, 1597, 2584, 4181, 6765, 10946, 17711, 28657]
fibs = np.array(fibs)
# logaritmos dos números com np.log()
# mas não pode dar erro...
logfibs = '???????'
plt.plot(np.arange(len(logfibs)), logfibs, 'o-')
plt.show()

Solução
Show code cell content
import numpy as np
from matplotlib import pyplot as plt
fibs = [0, 1, 1, 2, 3, 5, 8, 13, 21, 34, 55, 89, 144, 233, 377, 610, 987, 1597, 2584, 4181, 6765, 10946, 17711, 28657]
fibs = np.array(fibs)
# logaritmos dos números com np.log()
# mas não pode dar erro...
logfibs = np.log(fibs[fibs!=0])
plt.plot(np.arange(len(logfibs)), logfibs, 'o-')
plt.show()

11.2.3. Programa 3#
Sugestão: use a função np.log10() e a função np.trunc()
fibs = [1, 1, 2, 3, 5, 8, 13, 21, 34, 55, 89, 144, 233, 377, 610, 987, 1597, 2584, 4181, 6765, 10946, 17711, 28657]
fibs = np.array(fibs)
ndigits = '??????'
print('numero de dígitos:')
print(ndigits)
numero de dígitos:
[1. 1. 1. 1. 1. 1. 2. 2. 2. 2. 2. 3. 3. 3. 3. 3. 4. 4. 4. 4. 5. 5. 5.]
Solução
Show code cell content
fibs = [1, 1, 2, 3, 5, 8, 13, 21, 34, 55, 89, 144, 233, 377, 610, 987, 1597, 2584, 4181, 6765, 10946, 17711, 28657]
fibs = np.array(fibs)
ndigits = np.trunc(1 + np.log10(fibs))
print('numero de dígitos:')
print(ndigits)
numero de dígitos:
[1. 1. 1. 1. 1. 1. 2. 2. 2. 2. 2. 3. 3. 3. 3. 3. 4. 4. 4. 4. 5. 5. 5.]
11.3. Sobre processamento de ficheiros de texto estruturados#
11.3.1. Preparação#
Obtenha do moodle o ficheiro proteins.fasta e coloque na mesma pasta que o “programa 4”.
11.3.2. Programa 4#
with open('proteins.fasta') as f:
records = f.read().split('>')
# clean empty records
records = [p for p in records if len(p) > 0]
print(len(records))
headers = '??????'
descriptions = []
accs = []
# header example:
# sp|Q08641|AB140_YEAST tRNA(Thr) (cytosine(32)-N(3))-methyltransferase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) ...
for header in headers:
sp, acc, rest = header.split('|')
description = rest.split('??????')[0]
accs.append(acc)
descriptions.append(description)
for h, d in zip(accs, descriptions):
print(h, d)
4
P38090 AGP2_YEAST General amino acid permease AGP2
Q12001 ALG6_YEAST Dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase
P53309 AP18B_YEAST Clathrin coat assembly protein AP180B
P40467 ASG1_YEAST Activator of stress genes 1
Solução
Show code cell content
with open('proteins.fasta') as f:
records = f.read().split('>')
# clean empty records
records = [p for p in records if p]
print(len(records))
headers = [record.splitlines()[0] for record in records]
descriptions = []
accs = []
# header example:
# sp|Q08641|AB140_YEAST tRNA(Thr) (cytosine(32)-N(3))-methyltransferase OS=Saccharomyces cerevisiae ...
for header in headers:
sp, acc, rest = header.split('|')
description = rest.split('OS=')[0]
accs.append(acc)
descriptions.append(description)
for h, d in zip(accs, descriptions):
print(h, d)
4
P38090 AGP2_YEAST General amino acid permease AGP2
Q12001 ALG6_YEAST Dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase
P53309 AP18B_YEAST Clathrin coat assembly protein AP180B
P40467 ASG1_YEAST Activator of stress genes 1
